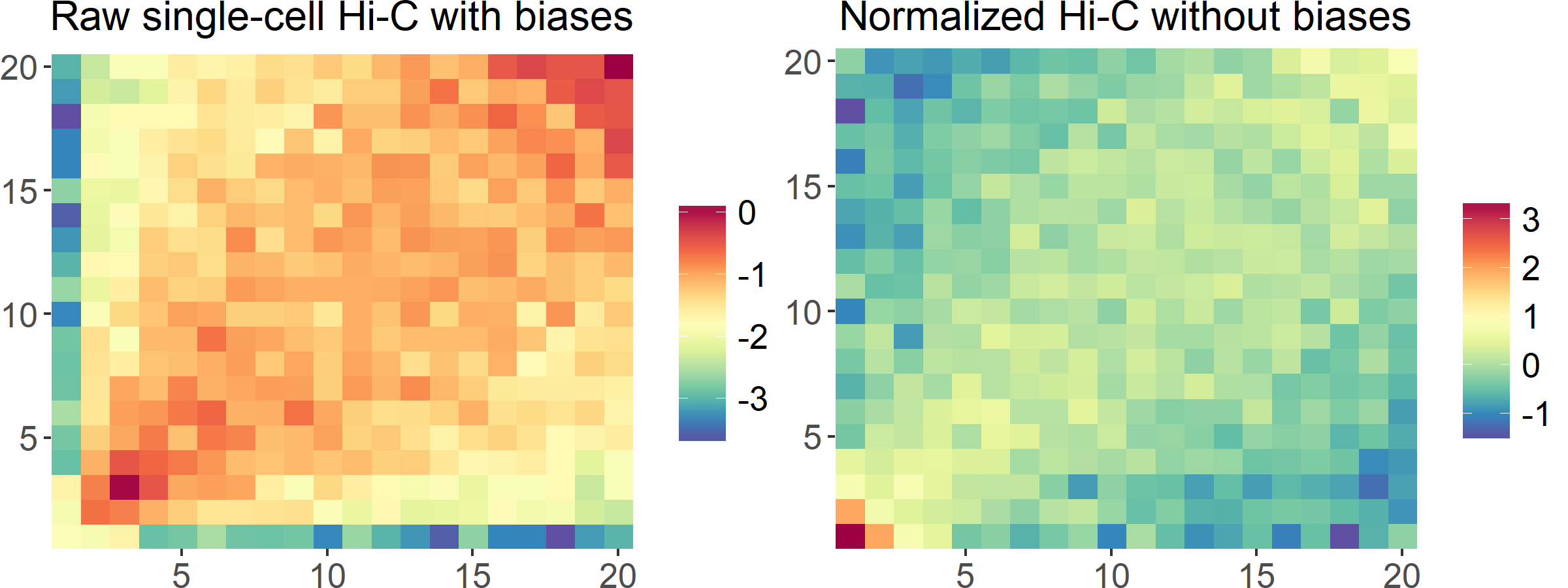
Introduction to scHiCNorm
scHiCNorm is a software package for eliminating systematic biases in single-cell Hi-C data. Considering that single-cell Hi-C data are zero-inflated, here we use zero-inflated (Poisson and Negative Binomial) and hurdle (Poisson and Negative Binomial) models to remove biases, including cutting sites, GC content, and mappability.
Download
Source code can be downloaded here (16K). And you can get the feature files (DpnII, cutting enzyme) for Human-hg19 (620M), Human-hg38 (622M), Mouse-mm9 (544M), and Mouse-mm10 (545M). You can also download the feature files for a specific chromosome here.
Reference
Tong Liu and Zheng Wang. scHiCNorm: a software package to eliminate systematic biases in single-cell Hi-C data. Bioinformatics 2018;34(6):1046-1047.
Contact
For any questions or suggestions, please contact:
Dr. Zheng Wang
Department of Computer Science
University of Miami