3DChrom: Reconstructing three-dimensional structures of chromatins and TADs using Hi-C data and MDS methods
Help Page
Given a symmetric Hi-C contact matrix, 3DChrom can reconstruct three-dimensional (3D) structures of chromatins or topologically associating domains (TADs). The methods are described as follows:
- Converting Hi-C contacts into wish distances. We define two mutually exclusive Hi-C contact sets based on whether Hi-C contacts equal zero:
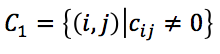 and
and 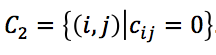 . The wish spatial distancs are obtained from the equaltion
. The wish spatial distancs are obtained from the equaltion 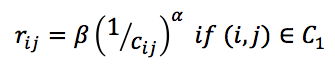 , where α is set to 1/3 and β is the scale parameter (default 1).
, where α is set to 1/3 and β is the scale parameter (default 1). - Defining objective functions to get the optimal coordinates in 3D space in accordance with the wish distancs. Here we provide three objective functions.
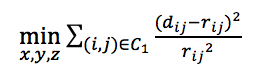 from PASTIS.
from PASTIS.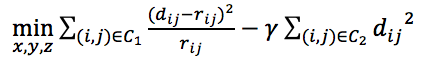 from ChromSDE.
from ChromSDE.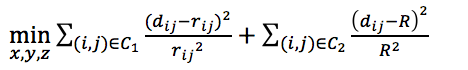 self-defined with R equal to maximum wish distance or using shortest path to infer the with distance of missing Hi-C contact.
self-defined with R equal to maximum wish distance or using shortest path to infer the with distance of missing Hi-C contact.- Obtaining optimal solutions (i.e., 3D coordinates) for the three objective functions using IPOPT.